A Biofizikai Önálló Osztály oldalai
Department of Biophysics
Whole-group meeting
In this semester our whole-group gathering is held every Wednesday from 10.00am where talks are given by invited guests and lab-members, and different topics are discussed usually in Hungarian. Before joining these meetings, please contact Kriszta Szalisznyó.
Computational Neuroscience Group, Department of Biophysics
The role of self-excitation in the development of topographic order
A connectionist type model is presented for explaining the development of the topographically ordered retinotectal connections of the visual system of the frog. The structure of a tectal column were taken into account inplicitly. Simulations suggest that the self-excitatory intracolumnar connections play an important role in the formation of the topography.
Computational Neuroscience Group, Department of Biophysics
Olfaction and its underlying stochastic phenomena
Computational Neuroscience Group, Department of Biophysics
Location dependent differences between somatic and dendritic IPSPs
Computational Neuroscience Group, Department of Biophysics
Development of hippocampal place fields
Computational Neuroscience Group, Department of Biophysics
A populational model of hippocampus CA3 region slices
A statistical theory of interacting neural fields, motivated by Ventriglia's kinetic theory is formulated.
Ventriglia, F. Kinetic approach to neural systems I. Bull. Math. Biol. 36:534--544. (1974)
Ventriglia, F. Computational simulation of activity of cortical-like neural systems. Bull. Math. Biol. 50:143--185 (1988)
Computational Neuroscience Group, Department of Biophysics
Generation and control of septo-hippocampal oscillations
Computational Neuroscience Group, Department of Biophysics
ICEA - Related publications
Recent hippocampus related publications
Place cell activity
- Lengyel M, Szatmáry Z, Érdi P.: Dynamically detuned oscillations account for the coupled rate and temporal code of place cell firing.
Computational Neuroscience Group, Department of Biophysics
ICEA - Cooperators
| Partner | Key person | Activity |
|---|---|---|
| SCAI Lab (project team), School of Humanities & Informatics, University of Skövde HIS | Tom Ziemke | A1-WP1,WP2,WP3 |
Computational Neuroscience Group, Department of Biophysics
ICEA - Activities and Workpackages
Activity 1 - Theoretical framework
- WP1 - initial theoretical framework
- WP2 - integration of the different research activtities
- WP3 - promotion of project results
Activity 2 - ICEAbot: an integrated robot platform
Computational Neuroscience Group, Department of Biophysics
ICEA - Modelling goal-directed navigation of the rat
The Consortium
Computational Neuroscience Group, Department of Biophysics
EURESIST - Integration of viral genomics with clinical data to predict response to anti-HIV treatment
Magyar nyelvű információk az EuResist projekttel kapcsolatban, nem szakemberek számára.
Purpose of the project
Computational Neuroscience Group, Department of Biophysics
Prediction of technological development from patent citation network
Computational Neuroscience Group, Department of Biophysics
Organisation of signal flow in directed networks
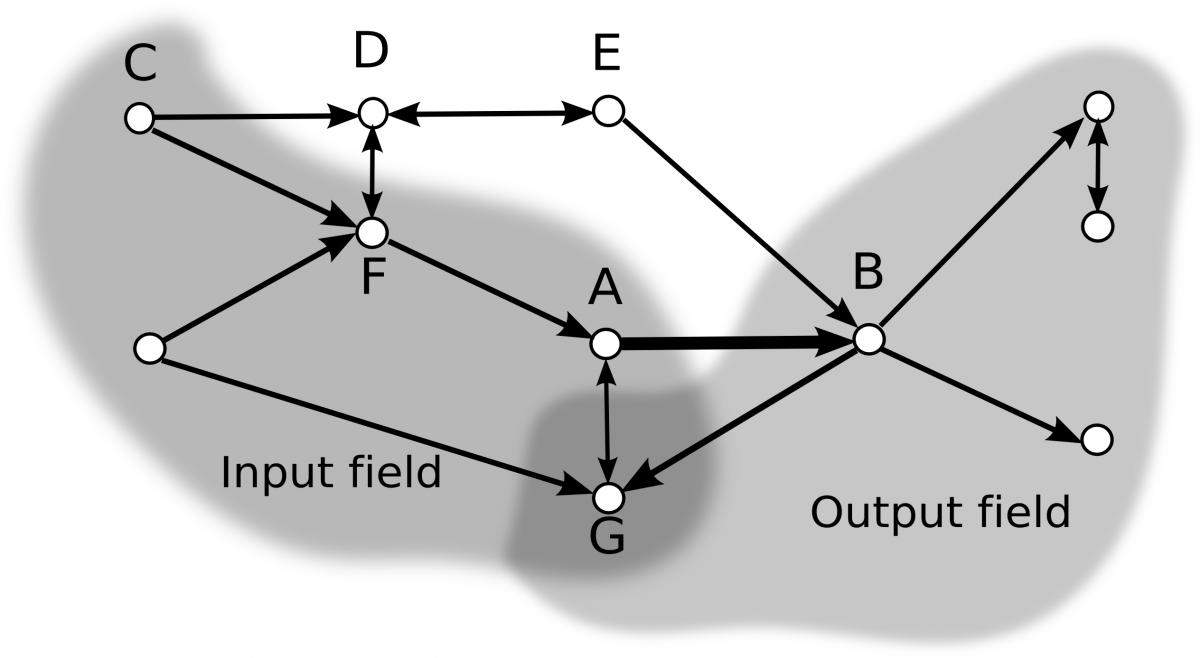
Computational Neuroscience Group, Department of Biophysics
Modeling of budget time series
Computational Neuroscience Group, Department of Biophysics
Software package for complex network analysis
The igraph library
The igraph library is a collection of functions for creating and manipulating graphs. It's main design goal is efficiency. It has started as an additional package to the GNU R statistical environment, but now most of its functionality is written in C and is available as a separate library.
